The counting of colonies to quantitate bacteria is an important process in labs for research, clinical diagnosis, and environmental monitoring e.g. in water. Generally counted manually, progress in technology has automated the process through colony counters that can be purchased but can be costly. While efficient, there remains also a challenge correctly detecting clustered colonies compromising inaccurate counts. Leveraging on the smartphone camera, we developed an app that processes images of colony plates and automatically counts them. To provide more accurate counts, the watershed algorithm with distance transform was incorporated into the segmentation and found to distinguish clustered or partially merged colonies better. This app, the APD Colony Counter app, thus transforms the smartphone into a portable colony counter, making the once laborious and tedious process manual colony counting, more accurate, automated and on-the-go.
Quantitating bacteria through colony counting on plates is routine in many biomedical, clinical, and environmental labs. In research, it can be used to determine the efficiency of lab processes and reagents e.g. competent cells (Chan et al., 2013), for determining quantifying bacteria in disease states (e.g. on skin wounds, see review by Kallstrom G, 2014), and environmental monitoring, such as in potable water (Reasoner DJ & Geldreich EE, 1985) and food (Delbès, C et al., 2007). Though a routine process, the manual counting of colonies is highly tedious and error-prone. Common manual counting practices include using a marker to dot counted colonies while counting and can take up to hours for plates with numerous partially merged colonies. To automate this, colony counters with image processing software have been released commercially. Costing from thousands of US dollars, these devices can scan culture plates to detect the number of colonies and produce a count in seconds. While the image processing software of these devices certainly made colony counting easier, there is still difficulty in segregating partially merged or clustered colonies. This often result in under reporting, which depending on the nature of the application, can have safety (e.g. food standards), medical (clinical standards) and environmental (e.g. natural water sources) consequences.
At present, each colony is assumed to arise from a single bacterium or colony-forming unit (CFU), thus when the colonies are clustered together or partially merged, inaccurate counts results in underestimation of the microbial load.
To increase the convenience of counting tasks, improve portability and accuracy in automated colony detection, the APD Colony Counter App was made to leverage on the smartphone and the in-built camera. Smartphones have been increasingly shown to be useful in biomedical research, replacing machines (e.g. GelApp, See Sim et al., 2015) and increasing the convenience of routine research procedures (see review by Gan SKE & Poon JK, 2016). In addition, they are increasingly making themselves felt in clinical use for diagnosis and patient care (Gan SKE et al., 2016). Colony counting apps does away with the need for costly equipment, enabling every smartphone owner to have a portable personalized counter. With the portability of the smartphone, the counter can be brought to the plate than vice versa, reducing contamination, and also facilitate automation. While there are many colony counter apps, the use of Itseez’s OpenCV http://opencv.org/ threshold and watershed algorithm with distance transform enables APD Colony Counter App to segregate merged colonies better for more accurate quantification.
APD Colony Counter App for Android was developed using version 4.2 of the Eclipse Integrated Development Environment, Juno and open source libraries: Mike Ortiz’s TouchImageView https://github.com/MikeOrtiz/TouchImageViewfor the editing, zooming, and panning functions on gel images; Itseez’s OpenCV http://opencv.org/ for the threshold and watershed algorithm with distance transform.
The image pre-processing step is crucial for accurate detection. To compensate for environmental factors (e.g. lighting, background) during photo taking, parameters have been built into the app such as brightness to intensify the colony objects against the background, and cropping to isolate only the area of interest to reduce interferences in the detection process.
Following image pre-processing, users can next adjust the threshold value of the image to achieve maximum contrast in getting white colonies on a black background for optimal automated detection. In the event that an image with black colonies on a white background is obtained, the “inverse image” button will invert the colors to give white colonies on a black background (Figure 1). With these parameters, the maximum contrast can be obtained for a variety of agar plates with different colors e.g. blood agar, where the colony and background agar colors vary.

Figure 1: Image with white colonies in black background after threshold adjustments.
Once these parameters are adjusted to a desired level, users can proceed to use the automated detection function to detect colonies. During this process, watershed segmentation with distance transform has been automatically incorporated to segregate merged colonies. The watershed algorithm separates merged colonies by internally placing lines to segregate distinct colonies.
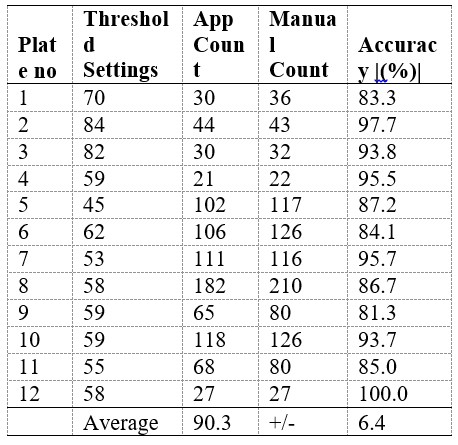
Table 1: Comparison of app and manually counted regions.
Data was obtained from 12 Agar plates.
As current image processing technologies are not able to achieve 100% accuracy ( Table 1) and automated detections are affected by image quality and filter settings, users can proceed to the edit interface to add and remove tagged colonies to correct undetected or falsely detected colonies.
For better visualization and inclusion in reports, the tags generated by APD Colony Counter can be modified for better visualization. Users can change the color, radius and thickness of the tags using a slide bar (Figure 2).
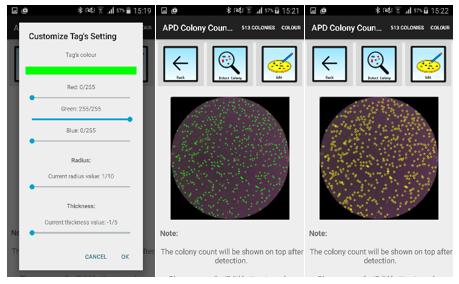
Figure 2: Tag Customization setting (left), default tags (middle) and tags with yellow scheme, increased radius and thickness value
For convenience, users can save their preferences of the tag visual effects by checking the ‘save settings’ box for future use.
APD Colony Counter App is a standalone Android application that functions as a pocket colony counter. Using the Watershed algorithm, it has the added advantage of segregating merged colonies better. With range of other image processing parameters, it can be used on a variety of agars to count colonies. Leveraging on the smartphone camera, it enable the smartphone to transform into a useful scientific device for the quantification of bacterial load in clinic, research, environmental, and even food safety regulatory labs.
Availability and implementation
The APD Colony Counter application is developed for the Android platform is freely available on the Google Play Store. More details on the app can be found at www.facebook.com/APDLab The APD Colony Counter user guide is available at http://tinyurl.com/ColonyApp, and a video tutorial is also available on the Google Play Store.
Contact: samuelg@bii.a-star.edu.sg; bii-apdlab@bii.a-star.edu.sg
Funding: This work is funded by the Joint Council Office, Agency for Science, Technology, and Research, Singapore
Chan WT, Verma CS, Lane DP, Gan SKE.(2013) A comparison and optimization of methods and factors affecting the transformation of Escherichia coli.Biosci. Rep. 33(6), art:e00086.doi:10.1042/BSR20130098.
Delbès, C., Ali-Mandjee, L., & Montel, M. C. (2007). Monitoring bacterial communities in raw milk and cheese by culture-dependent and-independent 16S rRNA gene-based analyses. Applied and environmental microbiology, 73(6), 1882-1891.
Gan SKE, Poon JK (2016) The World of Biomedical Apps: their uses, limitations, and potential. Scientific Phone Apps and Mobile Devices Vol 2:6. doi: 10.1186/s41070-016-0009-2.
Gan SKE, Koshy C, Nguyen PV, Haw YX (2016) An Overview of clinically and healthcare related apps in Google and Apple app stores: connecting patients, drugs, and clinicians. Scientific Phone Apps and Mobile Devices. Vol 2:8. doi: 10.1186/s41070-016-0012-7.
Kallstrom G (2014). Are Quantitative Bacterial Wound Cultures Useful? Journal of Clinical Microbiology. 2014 Aug; 52(8): 2753–2756. doi: 10.1128/JCM.00522-14
Reasoner, D. J., & Geldreich, E. E. (1985). A new medium for the enumeration and subculture of bacteria from potable water. Applied and environmental microbiology, 49(1), 1-7.
Sim JZ, Nguyen PV, Lee HK, Gan SKE. GelApp: Mobile gel electrophoresis analyzer. Nature Methods Application Notes. doi: 10.1038/an9643